The profiles of small molecule inhibitors differ when comparing in vitro and cellular potency
In 2018, we published a comparison between inhibitor profiles across the kinome generated using a biochemical in vitro assay or the cellular NanoBRETTM target engagement assay (Vasta, et al (2018)). We found the profiles for dasatinib and crizotinib differed tremendously!
What makes both assay formats different? In vitro assays frequently use only the catalytic domain of a purified enzyme as an input to determine enzyme activity. These proteins, that are most often expressed in bacterial cells or insect cells, are different from human expressed proteins e.g. based on their post-translational modifications. In the NanoBRETTM assay, we are using human HEK293T cells as an expression system for our targets as a full-length protein spanning not only the catalytic domain but also different domains that may be involved in scaffolding or protein structure and activity – thereby also creating human post-translational modification. In NanoBRETTM, we also take the cell membrane into account which your compounds need to pass in order to engage their target. In living cells, we also find endogenous levels of natural binding partners of our targets, e.g. Adenosine Triphosphate (ATP) at high concentrations of approximately 2 mM that will impact small molecule binding.
In our study from 2018, we found the individual protein kinases KM for ATP to be the reason for the differeing inhibitor profiles (and subsequently a shift in their respective EC50s compared to in vitro IC50s) – making clear that testing your small molecule in living cells is cruicial for understanding their impact on a cellular level!
in vitro
in cellulo
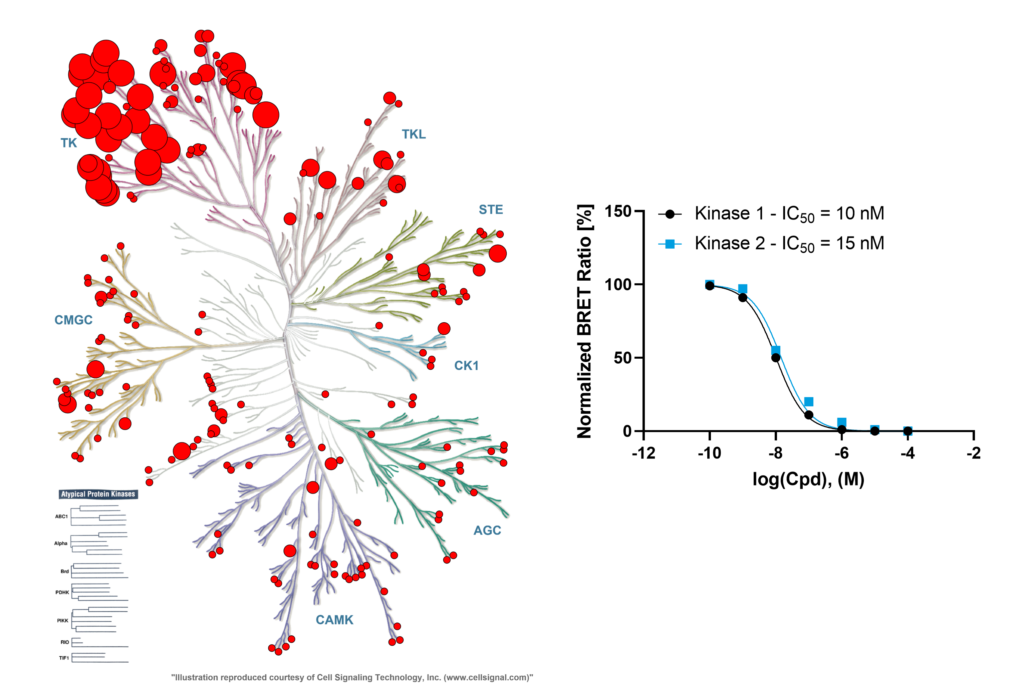
In vitro profile of dasatinib (Davis et al, 2011) shows many targets hit with high potency. Compound concentration was 1 µM, large cirlces indicate >91% binding, medium 50%, small 11%, very small <11%.
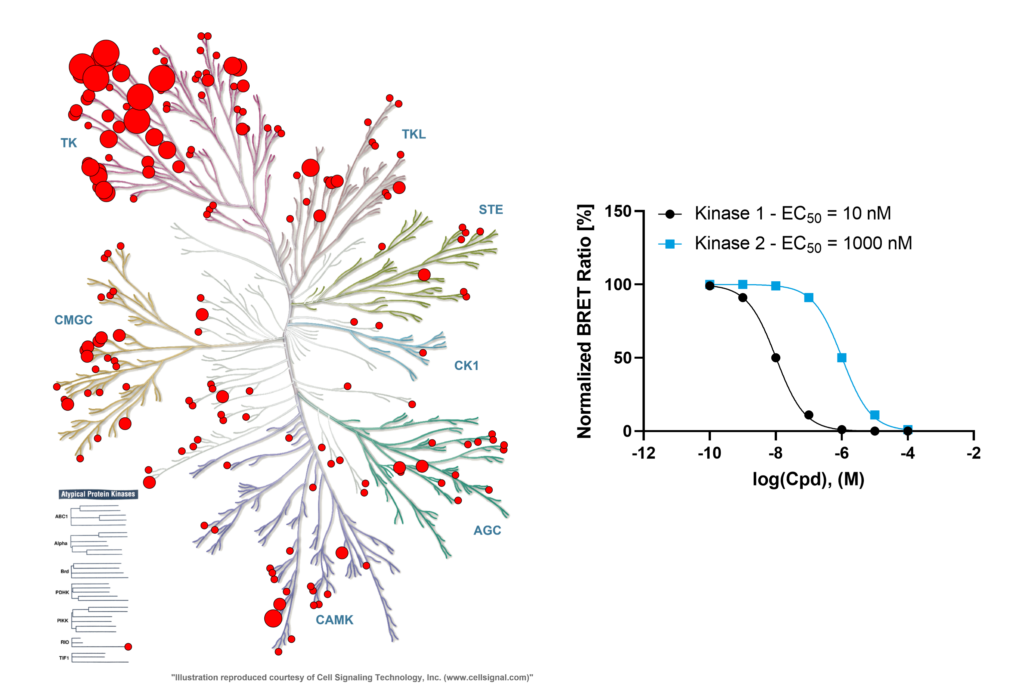
In cellulo profile of dasatinib (Vasta et al, 2018) shows less targets engaged. Compound concentration was 1 µM, large cirlces indicate >91% binding, medium 50%, small 11%, very small <11%.
- Purified enzyme
- Often catalytic domain only
- Often no post-translational modifications
- Measures protein inhibition
- No cell membrane
- ATP concentration at KM
- Transiently expressed protein
- Full-length protein
- Human post-translational modifications
- Measures protein binding
- Living cell with cell membrane
- Cellular concentration of ATP
NanoBRETTM is a test system we use to determine your inhibitors affinity in living cells.
NanoBRETTM is a cellular bioluminescence resonance energy transfer (BRET) based tracer competition assay developed by Promega. This means we use the light of the NLuc luciferase – a small 17 kDa protein that was optimized from a luciferase of the deep sea shrimp – as an energy donor and the BODIPY-based flourophore of our tracer molecule as a BRET acceptor if both are in close proximity. In the protein kinase target engagement assay, we transiently transfect living human HEK293T cells with our target of interest fused to a NLuc (provided by Promega). This leads to a mild overexpression of our target protein within overnight incubation time. Adding the (binding) tracer molecule will lead to a BRET between the NLuc as an energy donor and the BODIPY on the tracer as an energy acceptor upon luciferase substrate addition – a strong signal we can detect at a wavelength of 610 nm. We normalize the tracer signal by the luciferase signal at 450 nm creating a ratiometric assay that is independant of the expression of our target-luciferase complex or differences in cells dispensed or transfected in each experiment. If we now add a competitor – for example your small molecule of interest – the competitor will compete with the tracer thereby disrupting the BRET and subsequently reducing the BRET signal in a dose-dependent manner. If we plot this dose-dependent decrease in signal, we will obtain an EC50 for your competitor – a measure of affinity for your small molecule that can serve as a decision parameter within your set of small molecules you want to move forward with.
Because the NanoBRETTM is a competition assay, an excess of tracer molecule would prevent your competitor to compete with the tracer for the binding site. Hence, to make sure we can compare the EC50s obtained across different protein kinases, we always use the KD, app of the protein kinase-tracer complex that we determined in a separate experiment before.
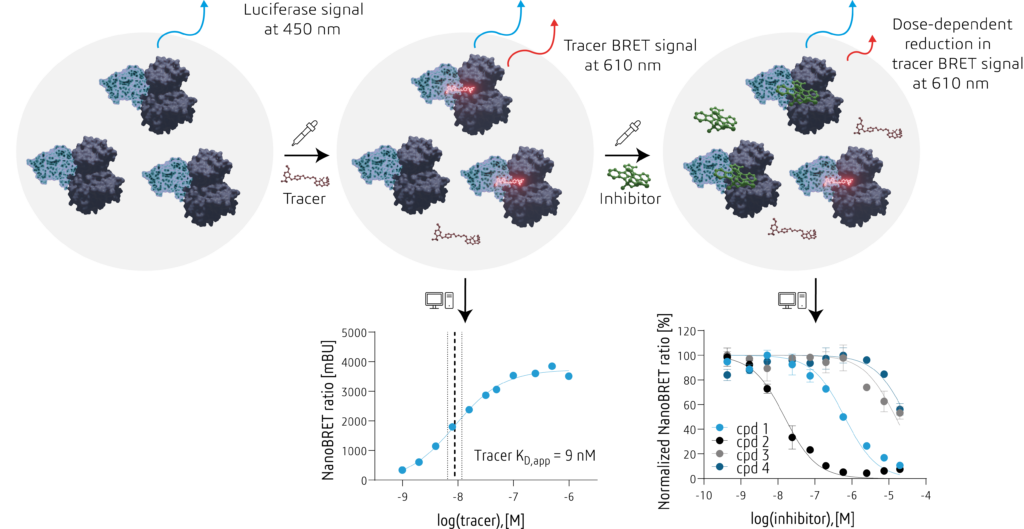
Using the NanoBRETTM technique we can answer many different questions. We can assess your small molecules potency via EC50 experiments, assess your inhibitors selectivity across the kinome in so called 1-shot assays at a single concentration or determine your inhibitors half-life in living cells. Please have a look at our different services for details.
If you are interested in either service or have any questions – please feel free to contact us!
